#load needed packages. make sure they are installed.
library(here) #for data loading/saving
library(dplyr)
library(skimr)
library(ggplot2)
library(tidyverse)
library(reshape2)Data Exploratory Analysis
Setup
Loading data
#Path to data.
data_location <- here::here("fluanalysis", "data", "cleaned_data.rds")
#load data
mydata <- readRDS(data_location)Looking at the data
dplyr::glimpse(mydata)Rows: 730
Columns: 32
$ SwollenLymphNodes <fct> Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, No, Yes, Y…
$ ChestCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y…
$ ChillsSweats <fct> No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, …
$ NasalCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y…
$ CoughYN <fct> Yes, Yes, No, Yes, No, Yes, Yes, Yes, Yes, Yes, No, …
$ Sneeze <fct> No, No, Yes, Yes, No, Yes, No, Yes, No, No, No, No, …
$ Fatigue <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye…
$ SubjectiveFever <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes…
$ Headache <fct> Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes…
$ Weakness <fct> Mild, Severe, Severe, Severe, Moderate, Moderate, Mi…
$ WeaknessYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye…
$ CoughIntensity <fct> Severe, Severe, Mild, Moderate, None, Moderate, Seve…
$ CoughYN2 <fct> Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes, Yes, Yes…
$ Myalgia <fct> Mild, Severe, Severe, Severe, Mild, Moderate, Mild, …
$ MyalgiaYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye…
$ RunnyNose <fct> No, No, Yes, Yes, No, No, Yes, Yes, Yes, Yes, No, No…
$ AbPain <fct> No, No, Yes, No, No, No, No, No, No, No, Yes, Yes, N…
$ ChestPain <fct> No, No, Yes, No, No, Yes, Yes, No, No, No, No, Yes, …
$ Diarrhea <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No,…
$ EyePn <fct> No, No, No, No, Yes, No, No, No, No, No, Yes, No, Ye…
$ Insomnia <fct> No, No, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Yes, Y…
$ ItchyEye <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes,…
$ Nausea <fct> No, No, Yes, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Y…
$ EarPn <fct> No, Yes, No, Yes, No, No, No, No, No, No, No, Yes, Y…
$ Hearing <fct> No, Yes, No, No, No, No, No, No, No, No, No, No, No,…
$ Pharyngitis <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, …
$ Breathless <fct> No, No, Yes, No, No, Yes, No, No, No, Yes, No, Yes, …
$ ToothPn <fct> No, No, Yes, No, No, No, No, No, Yes, No, No, Yes, N…
$ Vision <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, …
$ Vomit <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, N…
$ Wheeze <fct> No, No, No, Yes, No, Yes, No, No, No, No, No, Yes, N…
$ BodyTemp <dbl> 98.3, 100.4, 100.8, 98.8, 100.5, 98.4, 102.5, 98.4, …summary(mydata) SwollenLymphNodes ChestCongestion ChillsSweats NasalCongestion CoughYN
No :418 No :323 No :130 No :167 No : 75
Yes:312 Yes:407 Yes:600 Yes:563 Yes:655
Sneeze Fatigue SubjectiveFever Headache Weakness WeaknessYN
No :339 No : 64 No :230 No :115 None : 49 No : 49
Yes:391 Yes:666 Yes:500 Yes:615 Mild :223 Yes:681
Moderate:338
Severe :120
CoughIntensity CoughYN2 Myalgia MyalgiaYN RunnyNose AbPain
None : 47 No : 47 None : 79 No : 79 No :211 No :639
Mild :154 Yes:683 Mild :213 Yes:651 Yes:519 Yes: 91
Moderate:357 Moderate:325
Severe :172 Severe :113
ChestPain Diarrhea EyePn Insomnia ItchyEye Nausea EarPn
No :497 No :631 No :617 No :315 No :551 No :475 No :568
Yes:233 Yes: 99 Yes:113 Yes:415 Yes:179 Yes:255 Yes:162
Hearing Pharyngitis Breathless ToothPn Vision Vomit Wheeze
No :700 No :119 No :436 No :565 No :711 No :652 No :510
Yes: 30 Yes:611 Yes:294 Yes:165 Yes: 19 Yes: 78 Yes:220
BodyTemp
Min. : 97.20
1st Qu.: 98.20
Median : 98.50
Mean : 98.94
3rd Qu.: 99.30
Max. :103.10 head(mydata) SwollenLymphNodes ChestCongestion ChillsSweats NasalCongestion CoughYN Sneeze
1 Yes No No No Yes No
2 Yes Yes No Yes Yes No
3 Yes Yes Yes Yes No Yes
4 Yes Yes Yes Yes Yes Yes
5 Yes No Yes No No No
6 No No Yes No Yes Yes
Fatigue SubjectiveFever Headache Weakness WeaknessYN CoughIntensity CoughYN2
1 Yes Yes Yes Mild Yes Severe Yes
2 Yes Yes Yes Severe Yes Severe Yes
3 Yes Yes Yes Severe Yes Mild Yes
4 Yes Yes Yes Severe Yes Moderate Yes
5 Yes Yes Yes Moderate Yes None No
6 Yes Yes Yes Moderate Yes Moderate Yes
Myalgia MyalgiaYN RunnyNose AbPain ChestPain Diarrhea EyePn Insomnia
1 Mild Yes No No No No No No
2 Severe Yes No No No No No No
3 Severe Yes Yes Yes Yes No No Yes
4 Severe Yes Yes No No No No Yes
5 Mild Yes No No No No Yes Yes
6 Moderate Yes No No Yes Yes No No
ItchyEye Nausea EarPn Hearing Pharyngitis Breathless ToothPn Vision Vomit
1 No No No No Yes No No No No
2 No No Yes Yes Yes No No No No
3 No Yes No No Yes Yes Yes No No
4 No Yes Yes No Yes No No No No
5 No Yes No No Yes No No No No
6 No Yes No No Yes Yes No No No
Wheeze BodyTemp
1 No 98.3
2 No 100.4
3 No 100.8
4 Yes 98.8
5 No 100.5
6 Yes 98.4skimr::skim(mydata)| Name | mydata |
| Number of rows | 730 |
| Number of columns | 32 |
| _______________________ | |
| Column type frequency: | |
| factor | 31 |
| numeric | 1 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| SwollenLymphNodes | 0 | 1 | FALSE | 2 | No: 418, Yes: 312 |
| ChestCongestion | 0 | 1 | FALSE | 2 | Yes: 407, No: 323 |
| ChillsSweats | 0 | 1 | FALSE | 2 | Yes: 600, No: 130 |
| NasalCongestion | 0 | 1 | FALSE | 2 | Yes: 563, No: 167 |
| CoughYN | 0 | 1 | FALSE | 2 | Yes: 655, No: 75 |
| Sneeze | 0 | 1 | FALSE | 2 | Yes: 391, No: 339 |
| Fatigue | 0 | 1 | FALSE | 2 | Yes: 666, No: 64 |
| SubjectiveFever | 0 | 1 | FALSE | 2 | Yes: 500, No: 230 |
| Headache | 0 | 1 | FALSE | 2 | Yes: 615, No: 115 |
| Weakness | 0 | 1 | FALSE | 4 | Mod: 338, Mil: 223, Sev: 120, Non: 49 |
| WeaknessYN | 0 | 1 | FALSE | 2 | Yes: 681, No: 49 |
| CoughIntensity | 0 | 1 | FALSE | 4 | Mod: 357, Sev: 172, Mil: 154, Non: 47 |
| CoughYN2 | 0 | 1 | FALSE | 2 | Yes: 683, No: 47 |
| Myalgia | 0 | 1 | FALSE | 4 | Mod: 325, Mil: 213, Sev: 113, Non: 79 |
| MyalgiaYN | 0 | 1 | FALSE | 2 | Yes: 651, No: 79 |
| RunnyNose | 0 | 1 | FALSE | 2 | Yes: 519, No: 211 |
| AbPain | 0 | 1 | FALSE | 2 | No: 639, Yes: 91 |
| ChestPain | 0 | 1 | FALSE | 2 | No: 497, Yes: 233 |
| Diarrhea | 0 | 1 | FALSE | 2 | No: 631, Yes: 99 |
| EyePn | 0 | 1 | FALSE | 2 | No: 617, Yes: 113 |
| Insomnia | 0 | 1 | FALSE | 2 | Yes: 415, No: 315 |
| ItchyEye | 0 | 1 | FALSE | 2 | No: 551, Yes: 179 |
| Nausea | 0 | 1 | FALSE | 2 | No: 475, Yes: 255 |
| EarPn | 0 | 1 | FALSE | 2 | No: 568, Yes: 162 |
| Hearing | 0 | 1 | FALSE | 2 | No: 700, Yes: 30 |
| Pharyngitis | 0 | 1 | FALSE | 2 | Yes: 611, No: 119 |
| Breathless | 0 | 1 | FALSE | 2 | No: 436, Yes: 294 |
| ToothPn | 0 | 1 | FALSE | 2 | No: 565, Yes: 165 |
| Vision | 0 | 1 | FALSE | 2 | No: 711, Yes: 19 |
| Vomit | 0 | 1 | FALSE | 2 | No: 652, Yes: 78 |
| Wheeze | 0 | 1 | FALSE | 2 | No: 510, Yes: 220 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| BodyTemp | 0 | 1 | 98.94 | 1.2 | 97.2 | 98.2 | 98.5 | 99.3 | 103.1 | ▇▇▂▁▁ |
Checking variable names in the dataset
names(mydata) [1] "SwollenLymphNodes" "ChestCongestion" "ChillsSweats"
[4] "NasalCongestion" "CoughYN" "Sneeze"
[7] "Fatigue" "SubjectiveFever" "Headache"
[10] "Weakness" "WeaknessYN" "CoughIntensity"
[13] "CoughYN2" "Myalgia" "MyalgiaYN"
[16] "RunnyNose" "AbPain" "ChestPain"
[19] "Diarrhea" "EyePn" "Insomnia"
[22] "ItchyEye" "Nausea" "EarPn"
[25] "Hearing" "Pharyngitis" "Breathless"
[28] "ToothPn" "Vision" "Vomit"
[31] "Wheeze" "BodyTemp" Continuous outcome variable: Body Temperature
Looking at the distribution of body temperature, our continuous outcome variable
ggplot(mydata, aes(x = BodyTemp)) +
geom_histogram() + ggtitle("Distribution of Body Temperature") + theme_classic()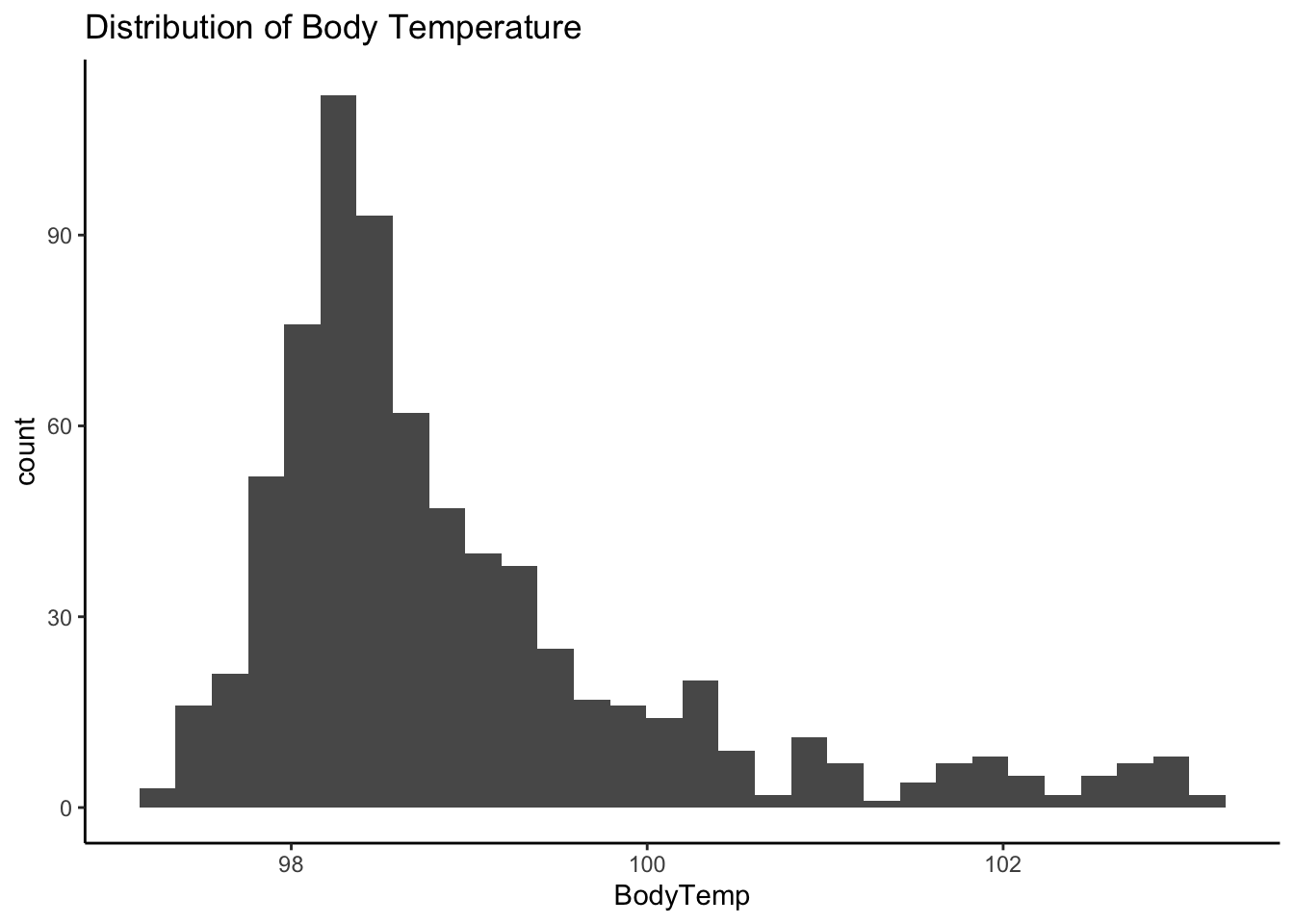
Creating boxplot of Cough intensity and Body temperature variables
# Creating boxplot of Cough intensity and Body temperature variables
ggplot(mydata, aes(x = factor(CoughIntensity), y = BodyTemp)) +
geom_boxplot() +
labs(x = "Cough Intensity", y = "Body Temperature") + ggtitle("Patterns between cough intensity abd body temperature") + theme_classic()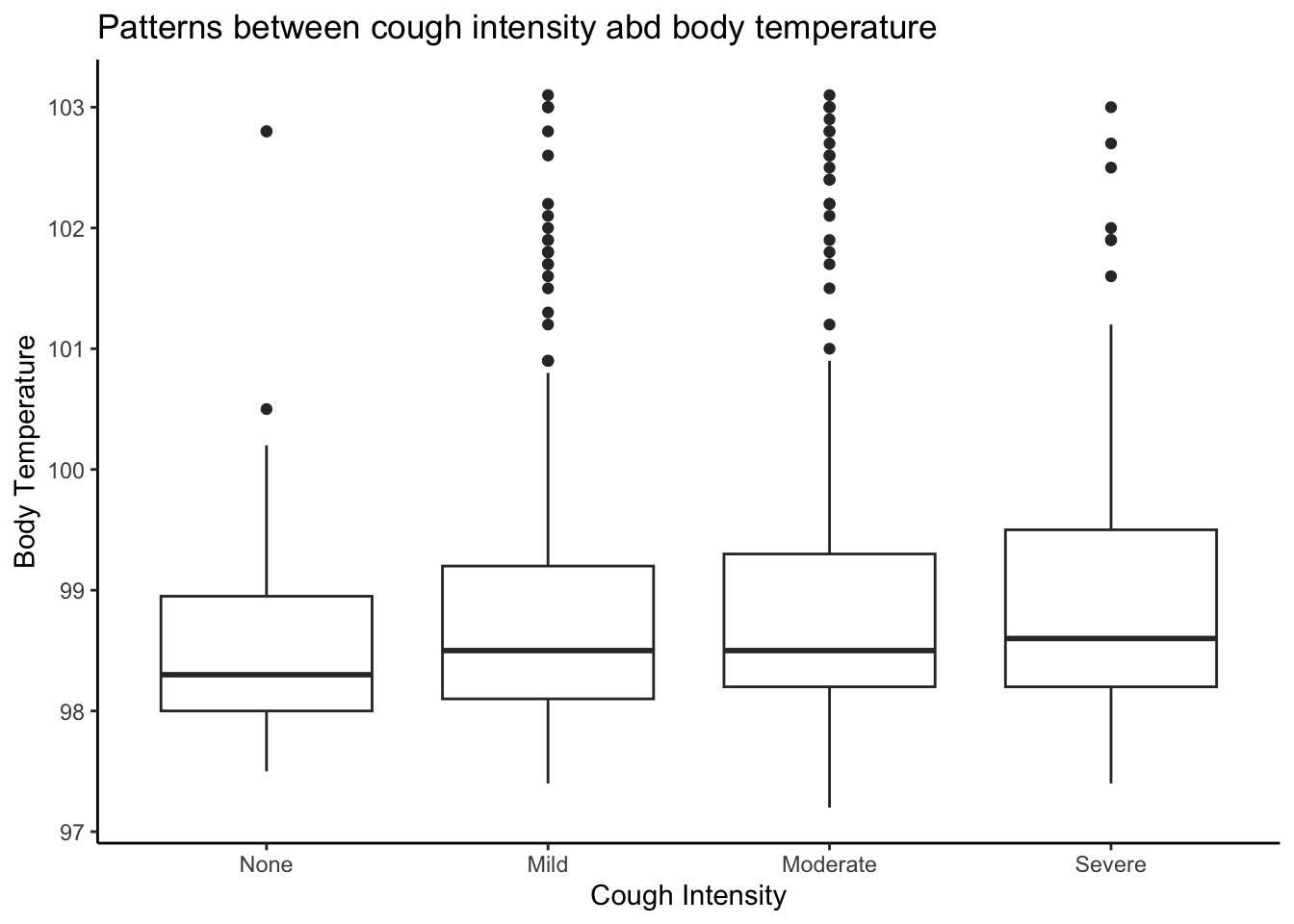
Creating boxplot of Chest Congestion and Body temperature variables
ggplot(mydata, aes(x = factor(ChestCongestion), y = BodyTemp)) +
geom_boxplot() +
labs(x = "Chest Congestion", y = "Body Temperature") + ggtitle("Patterns between ChestCongestion and body temperature") + theme_classic()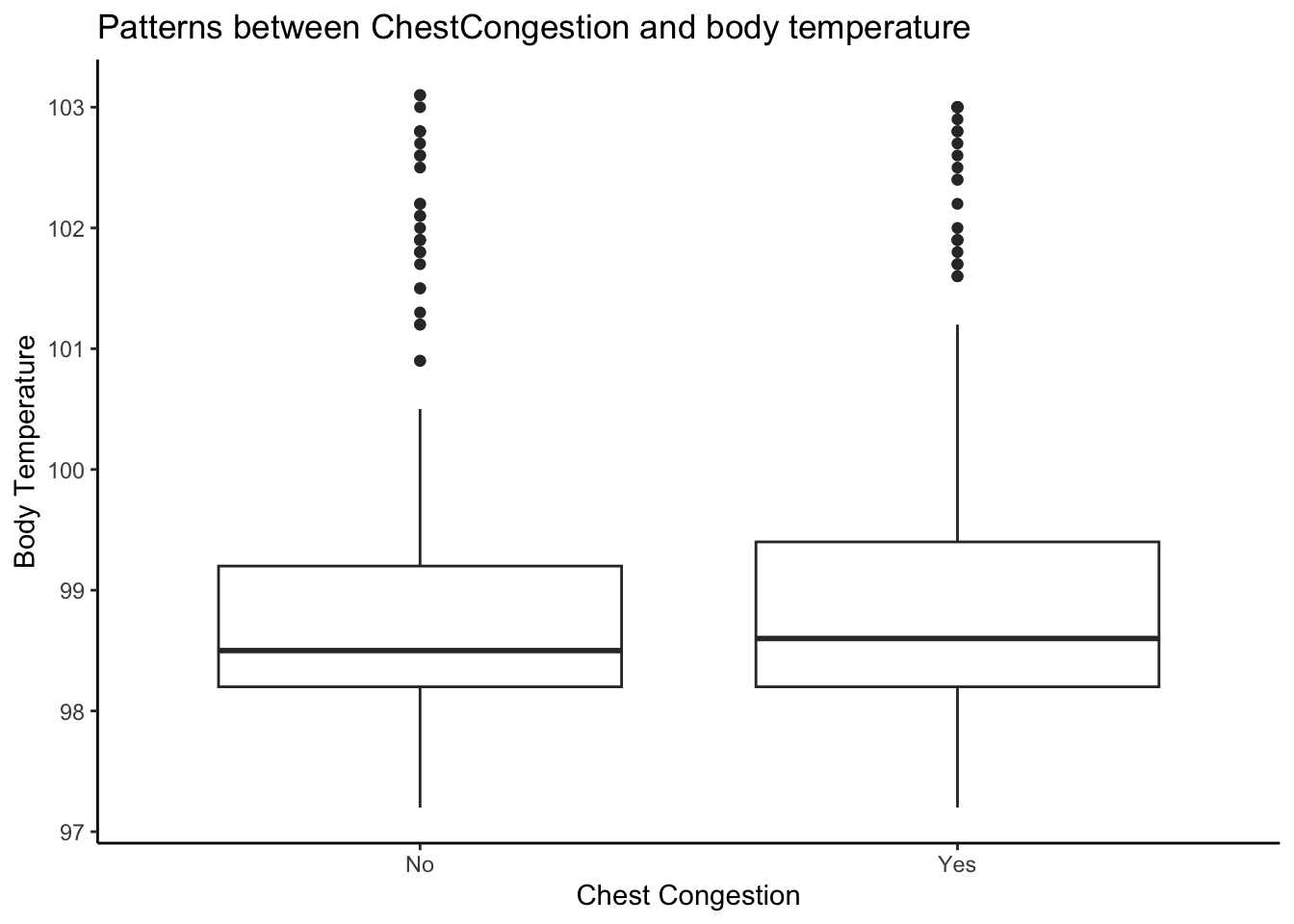
Creating boxplot of Headache and Body temperature variables
# Creating boxplot of Cough intensity and Body temperature variables
ggplot(mydata, aes(x = factor(Headache), y = BodyTemp)) +
geom_boxplot() +
labs(x = "Headache", y = "Body Temperature") + ggtitle("Patterns between Headache and body temperature") + theme_classic()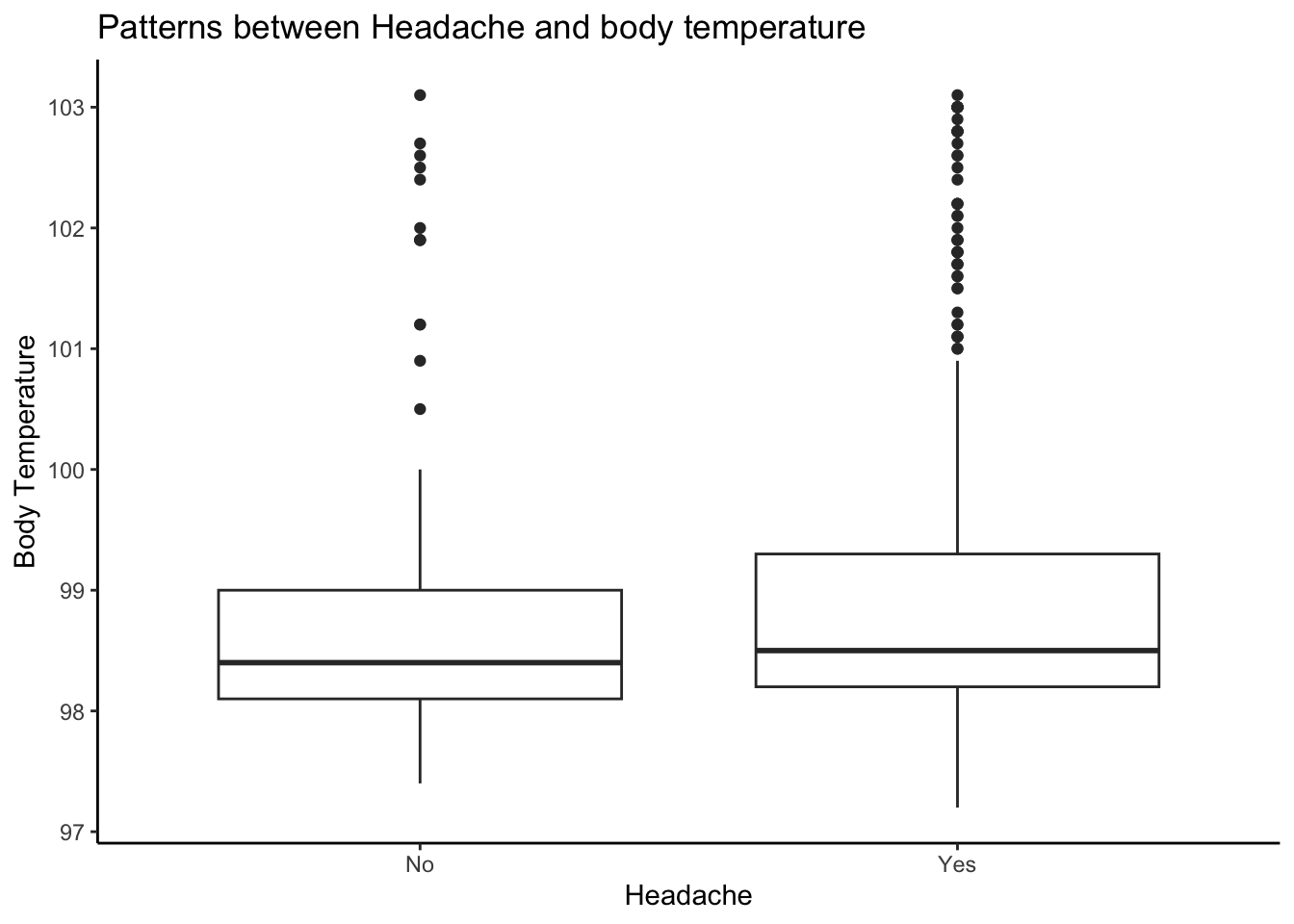
Categorical outcome variable
The distribution of Nausea
ggplot(mydata, aes(x = Nausea)) +
geom_bar() + ggtitle("Distribution of Nausea") + theme_classic()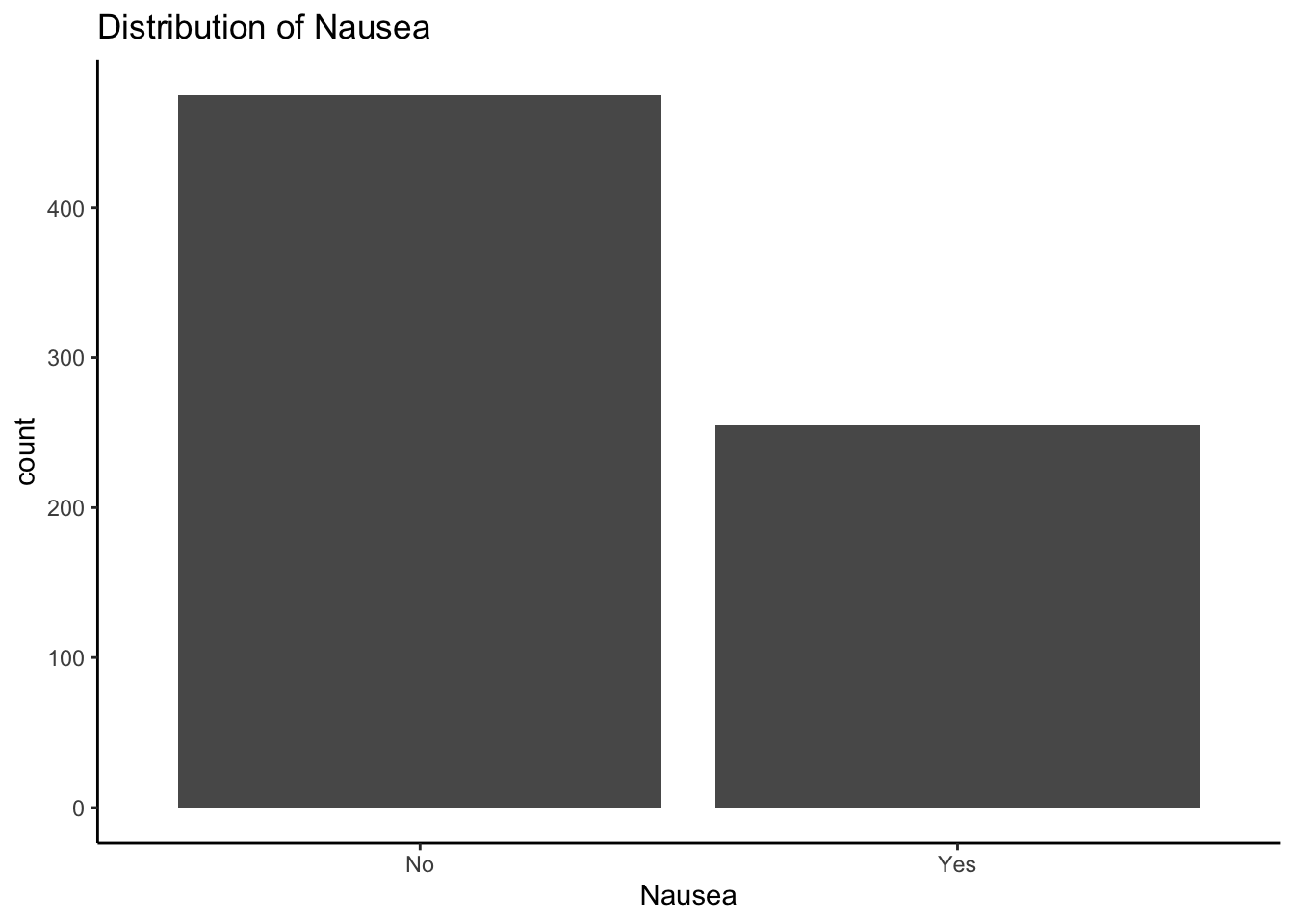
Distribution of Nausea and Nasal Congestion
ggplot(mydata, aes(x = Nausea, fill = NasalCongestion)) +
geom_histogram(stat="count") +
ggtitle("Distribution of Nausea and Nasal Congestion") + theme_classic()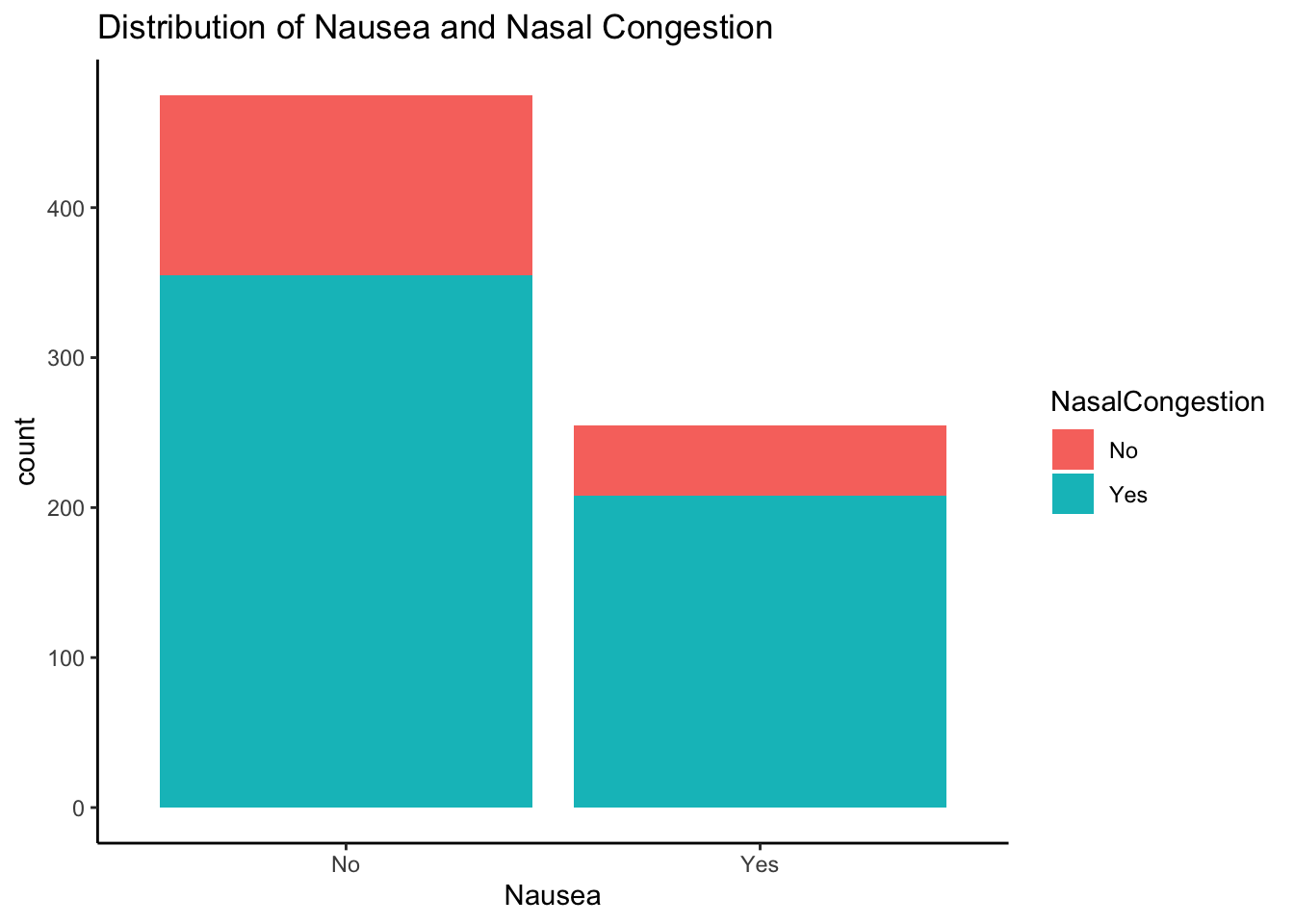
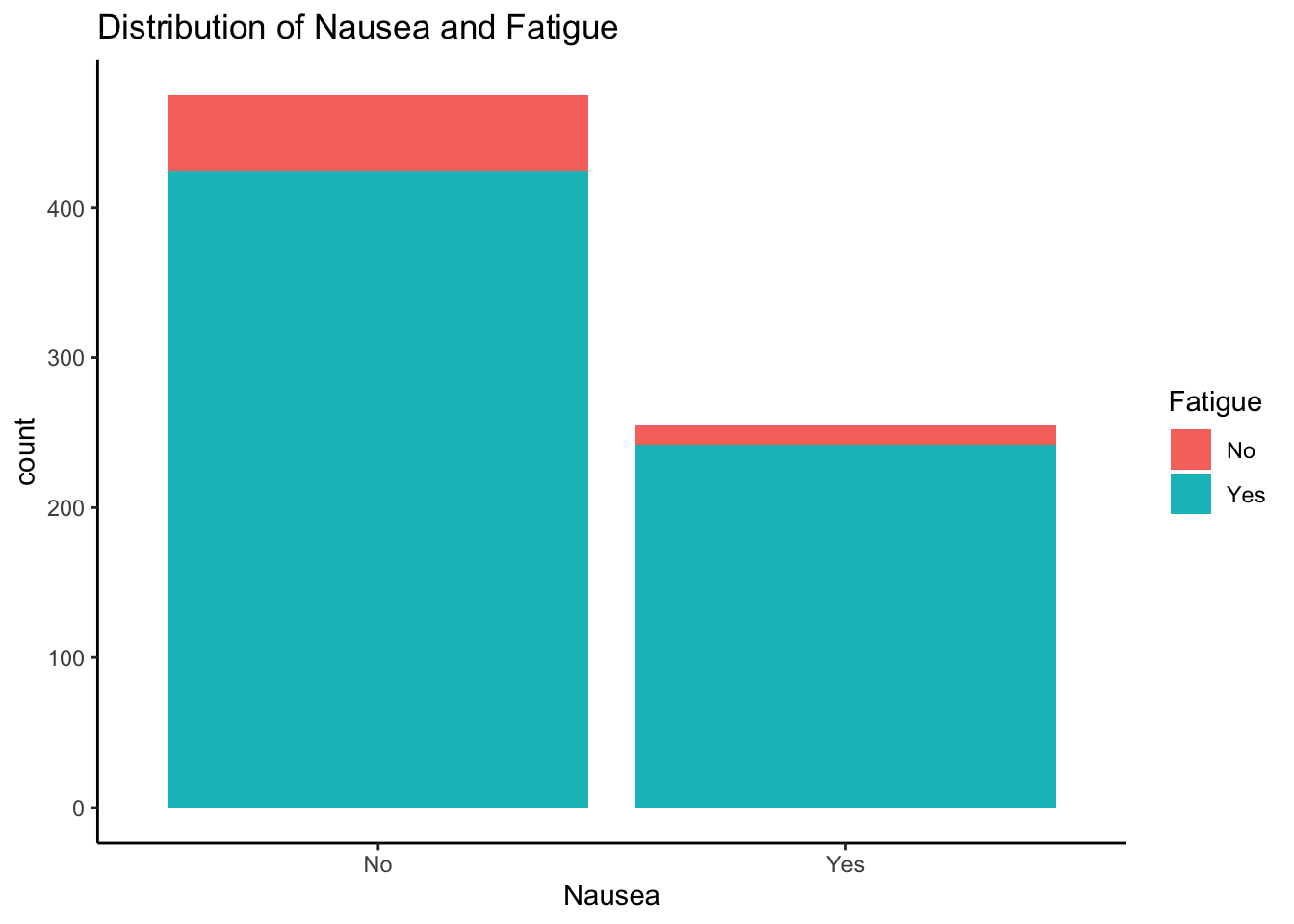
Distribution of Nausea and Nasal Congestion
ggplot(mydata, aes(x = Nausea, fill = Fatigue)) +
geom_histogram(stat="count") +
ggtitle("Distribution of Nausea and Fatigue") + theme_classic()