R Data Wrangling Exercise - Chris Okitondo
This dataset shows health conditions and contributing causes mentioned in conjunction with deaths involving coronavirus disease 2019 (COVID-19) by age group and jurisdiction of occurrence from 2020-2023. It was downloaded from the CDC website.
Setup
Load needed packages. make sure they are installed.
Data loading
# path to data
# note the use of the here() package and not absolute paths
data_location<- here::here("data","Conditions_Contributing_to_COVID-19_Deaths_2020_2023.xlsx")
rawdata <- readxl::read_excel(data_location)Looking at the data
dplyr::glimpse(rawdata)Rows: 521,640
Columns: 14
$ `Data As Of` <chr> "2023-01-29", "2023-01-29", "2023-01-29", "2023-0…
$ `Start Date` <chr> "2020-01-01", "2020-01-01", "2020-01-01", "2020-0…
$ `End Date` <chr> "2023-01-28", "2023-01-28", "2023-01-28", "2023-0…
$ Group <chr> "By Total", "By Total", "By Total", "By Total", "…
$ Year <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ Month <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ State <chr> "United States", "United States", "United States"…
$ `Condition Group` <chr> "Respiratory diseases", "Respiratory diseases", "…
$ Condition <chr> "Influenza and pneumonia", "Influenza and pneumon…
$ ICD10_codes <chr> "J09-J18", "J09-J18", "J09-J18", "J09-J18", "J09-…
$ `Age Group` <chr> "0-24", "25-34", "35-44", "45-54", "55-64", "65-7…
$ `COVID-19 Deaths` <dbl> 1492, 5700, 14878, 36982, 81436, 126144, 133948, …
$ `Number of Mentions` <dbl> 1562, 5916, 15491, 38430, 84400, 130069, 137110, …
$ Flag <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…summary(rawdata) Data As Of Start Date End Date Group
Length:521640 Length:521640 Length:521640 Length:521640
Class :character Class :character Class :character Class :character
Mode :character Mode :character Mode :character Mode :character
Year Month State Condition Group
Mode:logical Mode:logical Length:521640 Length:521640
TRUE:509220 TRUE:459540 Class :character Class :character
NA's:12420 NA's:62100 Mode :character Mode :character
Condition ICD10_codes Age Group COVID-19 Deaths
Length:521640 Length:521640 Length:521640 Min. : 0.0
Class :character Class :character Class :character 1st Qu.: 0.0
Mode :character Mode :character Mode :character Median : 0.0
Mean : 136.7
3rd Qu.: 21.0
Max. :1100296.0
NA's :153354
Number of Mentions Flag
Min. : 0.0 Length:521640
1st Qu.: 0.0 Class :character
Median : 0.0 Mode :character
Mean : 146.8
3rd Qu.: 23.0
Max. :1100296.0
NA's :148443 head(rawdata)# A tibble: 6 × 14
`Data As Of` Start D…¹ End D…² Group Year Month State Condi…³ Condi…⁴ ICD10…⁵
<chr> <chr> <chr> <chr> <lgl> <lgl> <chr> <chr> <chr> <chr>
1 2023-01-29 2020-01-… 2023-0… By T… NA NA Unit… Respir… Influe… J09-J18
2 2023-01-29 2020-01-… 2023-0… By T… NA NA Unit… Respir… Influe… J09-J18
3 2023-01-29 2020-01-… 2023-0… By T… NA NA Unit… Respir… Influe… J09-J18
4 2023-01-29 2020-01-… 2023-0… By T… NA NA Unit… Respir… Influe… J09-J18
5 2023-01-29 2020-01-… 2023-0… By T… NA NA Unit… Respir… Influe… J09-J18
6 2023-01-29 2020-01-… 2023-0… By T… NA NA Unit… Respir… Influe… J09-J18
# … with 4 more variables: `Age Group` <chr>, `COVID-19 Deaths` <dbl>,
# `Number of Mentions` <dbl>, Flag <chr>, and abbreviated variable names
# ¹`Start Date`, ²`End Date`, ³`Condition Group`, ⁴Condition, ⁵ICD10_codesskimr::skim(rawdata)| Name | rawdata |
| Number of rows | 521640 |
| Number of columns | 14 |
| _______________________ | |
| Column type frequency: | |
| character | 10 |
| logical | 2 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| Data As Of | 0 | 1.00 | 10 | 10 | 0 | 1 | 0 |
| Start Date | 0 | 1.00 | 10 | 10 | 0 | 37 | 0 |
| End Date | 0 | 1.00 | 10 | 10 | 0 | 37 | 0 |
| Group | 0 | 1.00 | 7 | 8 | 0 | 3 | 0 |
| State | 0 | 1.00 | 4 | 20 | 0 | 54 | 0 |
| Condition Group | 0 | 1.00 | 6 | 73 | 0 | 12 | 0 |
| Condition | 0 | 1.00 | 6 | 73 | 0 | 23 | 0 |
| ICD10_codes | 0 | 1.00 | 3 | 180 | 0 | 23 | 0 |
| Age Group | 0 | 1.00 | 3 | 10 | 0 | 10 | 0 |
| Flag | 368286 | 0.29 | 122 | 122 | 0 | 1 | 0 |
Variable type: logical
| skim_variable | n_missing | complete_rate | mean | count |
|---|---|---|---|---|
| Year | 12420 | 0.98 | 1 | TRU: 509220 |
| Month | 62100 | 0.88 | 1 | TRU: 459540 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 Deaths | 153354 | 0.71 | 136.69 | 3149.07 | 0 | 0 | 0 | 21 | 1100296 | ▇▁▁▁▁ |
| Number of Mentions | 148443 | 0.72 | 146.79 | 3376.53 | 0 | 0 | 0 | 23 | 1100296 | ▇▁▁▁▁ |
Cleaning
Telling R to avoid using scientific notation
options(scipen=999)List variable names
names(rawdata) [1] "Data As Of" "Start Date" "End Date"
[4] "Group" "Year" "Month"
[7] "State" "Condition Group" "Condition"
[10] "ICD10_codes" "Age Group" "COVID-19 Deaths"
[13] "Number of Mentions" "Flag" Let’s rename some variable names
rawdata <- rawdata %>% rename(current_data = `Data As Of`, start_date = `Start Date`, end_date = `End Date`, condition_group = `Condition Group`, condition = Condition, age_group = `Age Group`, covid_deaths = `COVID-19 Deaths`)Distribution of age group
table(rawdata$age_group)
0-24 25-34 35-44 45-54 55-64 65-74 75-84
52164 52164 52164 52164 52164 52164 52164
85+ All Ages Not stated
52164 52164 52164 Excluding data where age group is not stated
rawdata <- rawdata %>% filter(age_group != 'Not stated')Observation went from 521640 to 469476
Let’s visualize at COVID deaths by age group
ggplot(data = rawdata, aes(x = age_group, y = covid_deaths)) + geom_bar(stat='identity', fill = "steelblue")Warning: Removed 153109 rows containing missing values (`position_stack()`).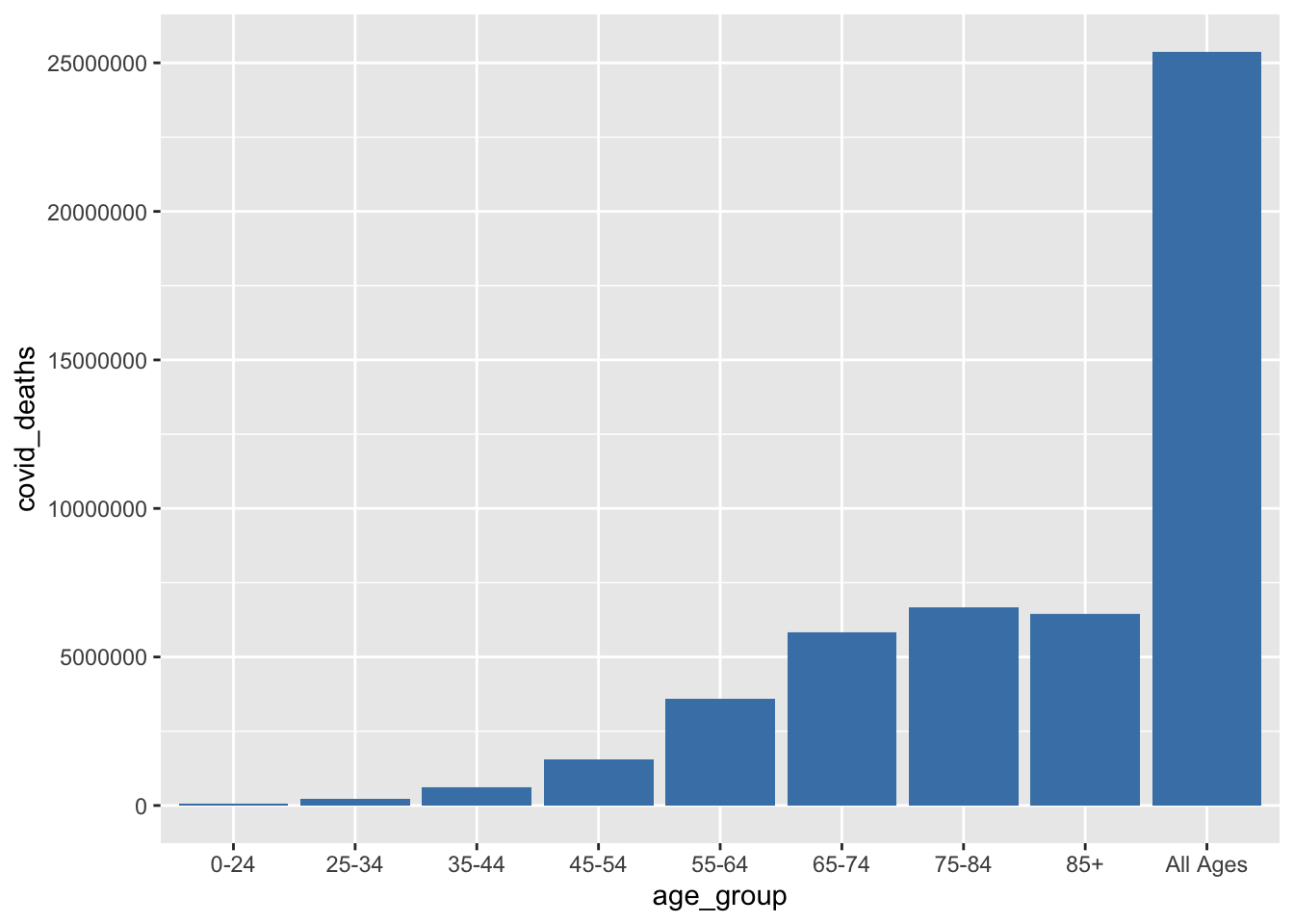
Saving the cleaned data
save_data_location <- here::here("data", "processeddata.rds")
saveRDS(rawdata, file = save_data_location)##This Section Added by Annabella Hines
cleandata <- readRDS(file=save_data_location)##Plots
First I wanted to look at the number of Covid deaths by each condition group.
cleandata2 <- cleandata %>% filter(Group=="By Total")
ggplot(cleandata2, aes(x=condition_group, y=covid_deaths))+geom_col()+xlab("Condition Group")+ylab("Covid Deaths")+ theme(axis.text.x = element_text(angle=13, size=5))Warning: Removed 1659 rows containing missing values (`position_stack()`).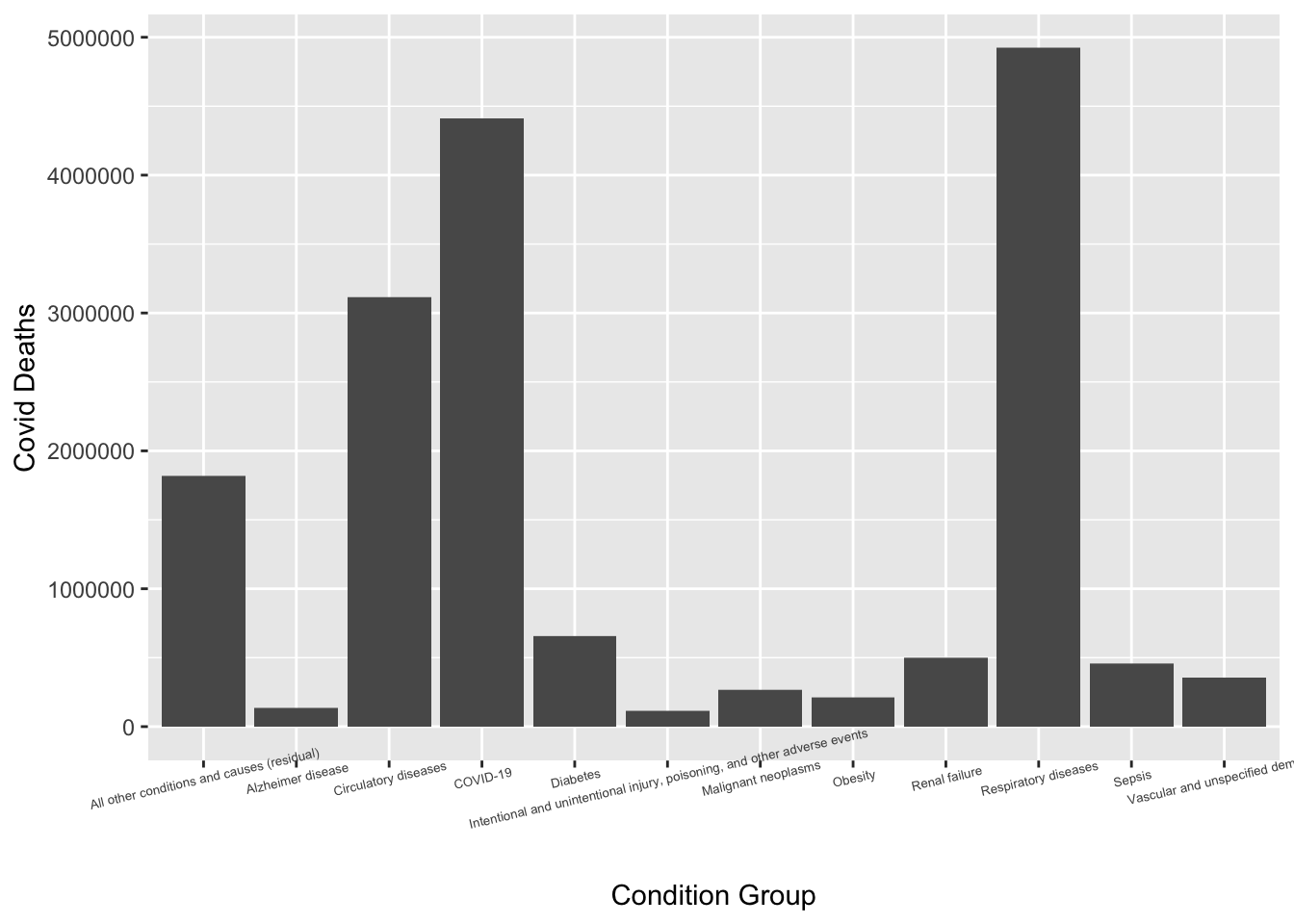
It appears that respiratory diseases had the highest number of associated Covid deaths. I now want to look at the respiratory disease by specific condition.
respiratory <- cleandata2 %>% filter(condition_group=="Respiratory diseases")
ggplot(respiratory, aes(x=condition, y=covid_deaths))+geom_col()+xlab("Respiratory Condition")+ylab("Covid Deaths")+theme(axis.text.x = element_text(angle=13))Warning: Removed 436 rows containing missing values (`position_stack()`).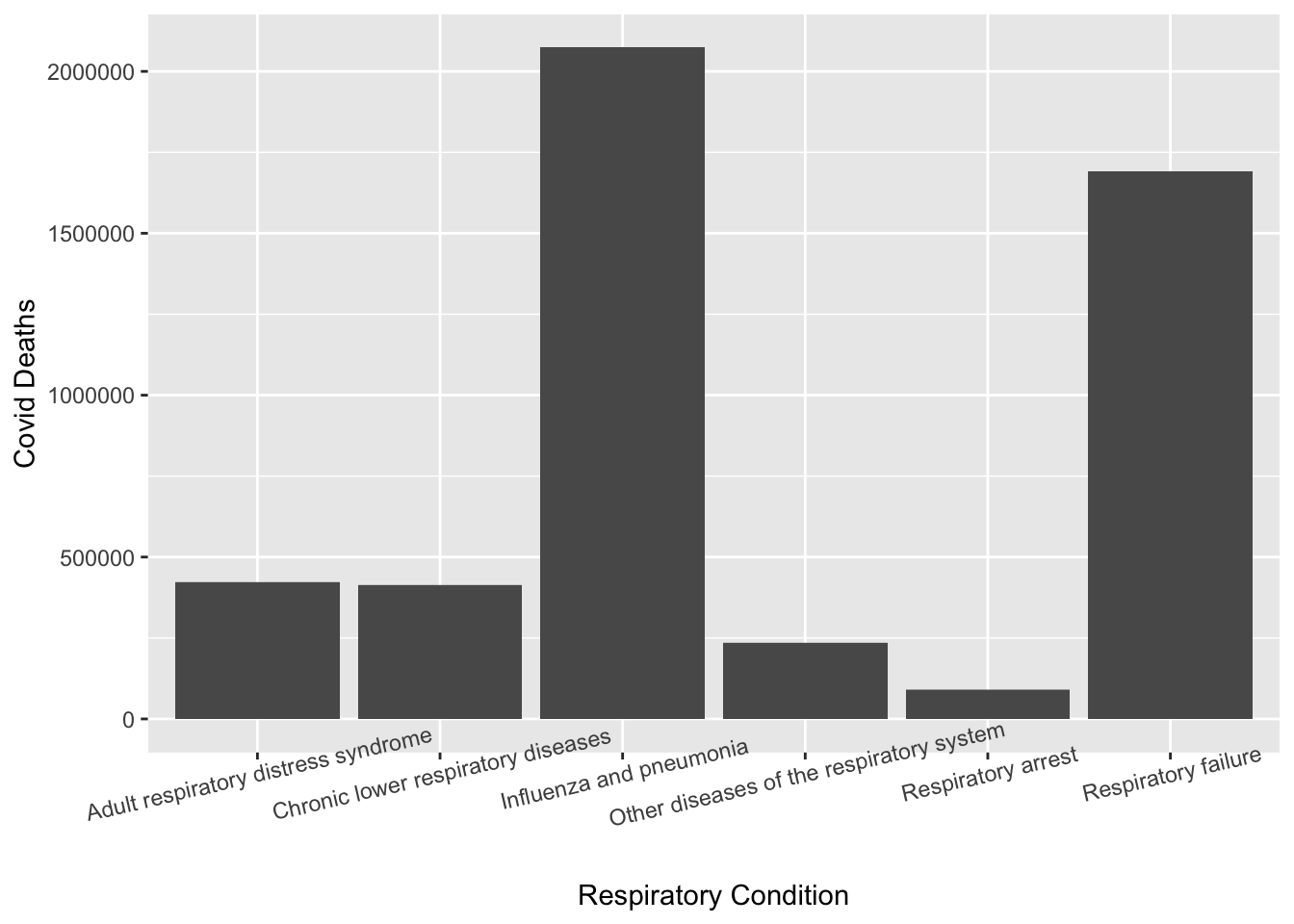
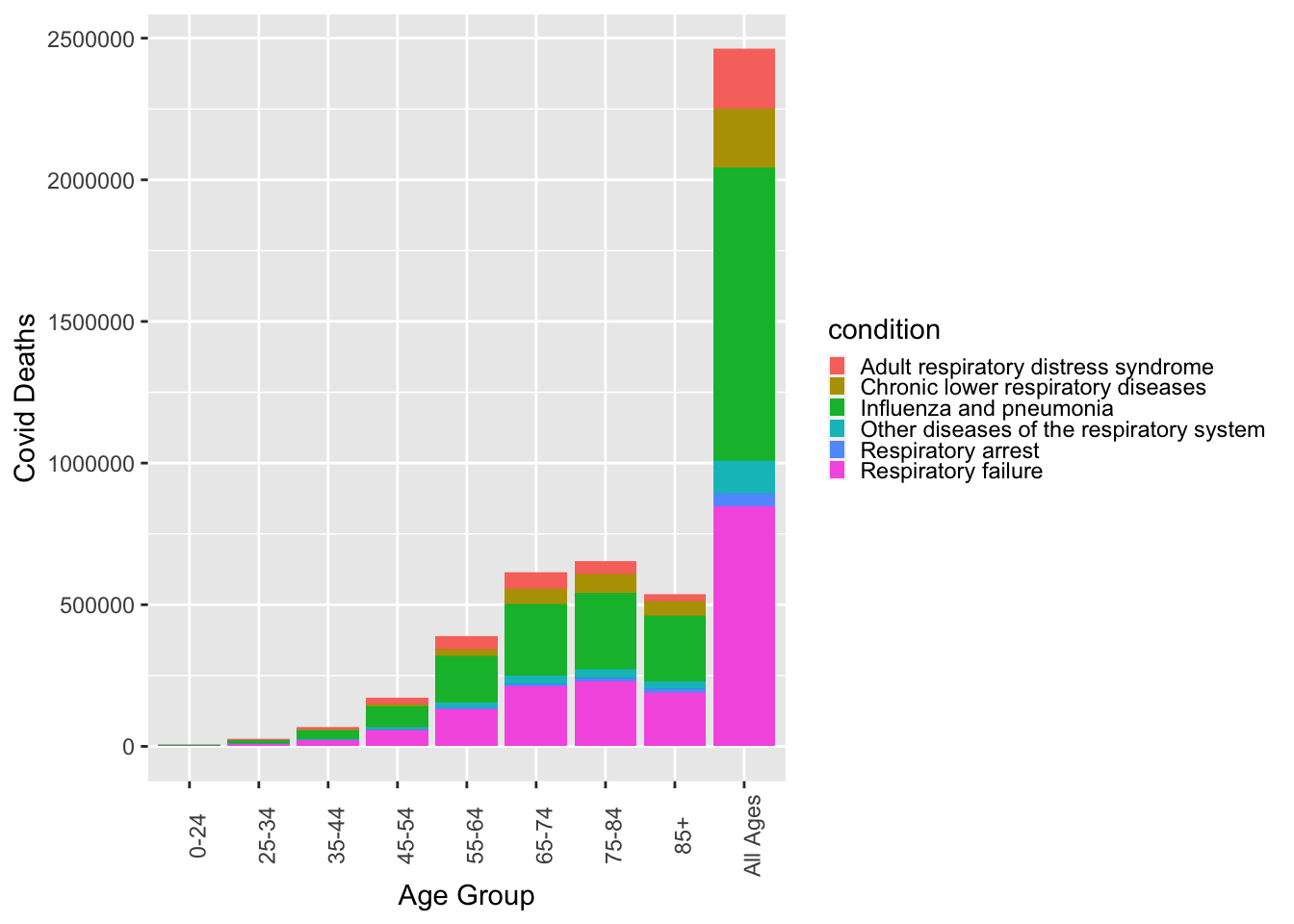
The respiratory condition with the highest number of Covid deaths appears to be the influenza and pneumonia. The last thing I want to do is look as the distribution of respiratory diseases by age group.
ggplot(respiratory, aes(x=age_group, y=covid_deaths, fill=condition))+geom_col()+theme(legend.key.size = unit(0.25, 'cm'), axis.text.x = element_text(angle=90))+ylab("Covid Deaths")+ xlab("Age Group")Warning: Removed 436 rows containing missing values (`position_stack()`).